No edit summary |
No edit summary |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{BioPsy}} |
{{BioPsy}} |
||
| + | {{Expert}} |
||
| − | In [[biochemistry]], a '''ligand''' is an [[effector (biology)|effector]], a [[molecule]] that binds to a site on a [[macromolecule]]'s surface by intermolecular forces, thereby changing the chemical conformation of the macromolecule. Once a molecule's conformation has changed, its ability to function in other chemical reactions is altered. This binding is usually a reversible reaction, i.e. it can be undone. Actual [[coordinate covalent bond]]s between a ligand and its target molecule are rare in biological systems. Ligands include [[substrate]]s, [[Enzyme inhibitor|inhibitor]]s, [[activator]]s, and [[neurotransmitter]]s. |
||
| + | [[Image:Myoglobin and heme.png|thumb|300px|right|[[Myoglobin]] (blue) with its ligand [[heme]] (orange) bound. Based on PDB ID 1MBO]] |
||
| + | '''Receptor binding''' or '''ligand binding''' is the process by which a [[ligand]] eg a neurotransmitter attaches itself to a [[receptor]] |
||
| − | Whether or not the ligand actually binds at a [[metal]] site is irrelevant, as opposed to '[[ligand]]' in chemical sense. 'Ligand' is probably originally a carryover term from the large number of binding studies on oxygen transport [[protein]]s, such as [[hemoglobin]], in which the ligand does indeed bind at a metal site. In the case of hemoglobin oxygen acts as a ligand. |
||
| + | . |
||
| + | In [[biochemistry]], a '''ligand''' (''latin'' ligare = to bind) is a [[Chemical substance|substance]] that is able to bind to and form a [[Complex (chemistry)|complex]] with a [[biomolecule]] to serve a biological purpose. Such binding is specific, as in [[neurochemistry]] where [[neurotransmitters]] bind only to specific [[receptors]] so [[dopamine]] will bind to a [[dopamine receptor]] and not to any other. |
||
| + | In a narrower sense, it is a signal triggering molecule binding to a [[binding site|site]] on a target [[protein]], by [[intermolecular force]]s such as [[ionic bond]]s, [[hydrogen bond]]s and [[Van der Waals force]]s. The docking (association) is usually reversible (dissociation). Actual irreversible [[covalent]] binding between a ligand and its target molecule is rare in biological systems. Ligand binding to [[Receptor (biochemistry)|receptor]]s alters the [[chemical conformation]], i.e. the three dimensional shape of the receptor protein. The conformational state of a receptor protein determines the functional state of a receptor. The tendency or strength of binding is called [[#Receptor/Ligand binding affinity|affinity]]. Ligands include [[Substrate (biochemistry)|substrate]]s, [[Enzyme inhibitor|inhibitor]]s, [[activator]]s, and [[neurotransmitter]]s. [[Radioligand]]s are [[radioisotope]] labeled compounds and used [[in vivo]] as [[tracer]]s in [[Positron emission tomography|PET]] studies and for [[in vitro]] [[binding study|binding studies]]. |
||
| − | [[Protein ligands]] are studied in [[structural biology]] and [[metabolomics]]. Radioactive ligands ([[radioligand]]s) are used together with [[positron emission tomography]] to study the [[receptor (biochemistry)|receptor]] systems of the brain. |
||
| − | |||
| − | In basic biology, ligands attach to receptors as a way of cytoplasmic signaling in a cell. The two main types of receptors that these ligands can attach to are soluble cytoplasmic proteins or [[transmembrane receptor]]s. An example of a cytoplasmic protein is a steroid receptor, where the ligand must be hydrophobic to get through the cell membrane, then attach to their desired protein within the cytoplasm. |
||
==Receptor/Ligand binding affinity== |
==Receptor/Ligand binding affinity== |
||
| Line 12: | Line 13: | ||
[[Image:Agonist.png|thumb|Two agonists with similar binding affinity]] |
[[Image:Agonist.png|thumb|Two agonists with similar binding affinity]] |
||
| − | A ligand that can bind to a receptor, alter the function of the receptor and trigger a physiological response is called an [[agonist]] for that receptor. Agonist binding to a receptor can be characterized both in terms of how much physiological response can be triggered and the [[concentration]] of the agonist that is required to produce the physiological response. High affinity ligand binding implies that a relatively low concentration of a ligand is adequate to maximally occupy a ligand binding site and trigger a physiological response. Low affinity binding implies that a relatively high concentration of a ligand is required before the binding site is maximally occupied and the maximum physiological response to the ligand is achieved. In the example shown to the right, two different ligands bind to the same receptor binding site. Only one of the agonists shown can maximally stimulate the receptor and |
+ | A ligand that can bind to a receptor, alter the function of the receptor and trigger a physiological response (eg the functioning of a neuron) is called an [[agonist]] for that receptor. Agonist binding to a receptor can be characterized both in terms of how much physiological response can be triggered and the [[concentration]] of the agonist that is required to produce the physiological response. High affinity ligand binding implies that a relatively low concentration of a ligand is adequate to maximally occupy a ligand binding site and trigger a physiological response. Low affinity binding implies that a relatively high concentration of a ligand is required before the binding site is maximally occupied and the maximum physiological response to the ligand is achieved. In the example shown to the right, two different ligands bind to the same receptor binding site. Only one of the agonists shown can maximally stimulate the receptor and thus can be defined as a "full agonist". An agonist that can only partially activate the physiological response is called a "partial agonist". Ligands that bind to a receptor but fail to activate the physiological response are receptor "[[Receptor antagonist|antagonists]]". In this example, the concentration at which the full agonist (red curve) can half-maximally activate the receptor is about 5 x 10<sup>-9</sup> [[Concentration#Molarity|Molar]] (nM = [[nano]]molar). |
[[Image:Agonists.png|thumb|left|400px|Two ligands with different receptor binding affinity.]] |
[[Image:Agonists.png|thumb|left|400px|Two ligands with different receptor binding affinity.]] |
||
| + | |||
| − | In the example shown to the left, ligand binding curves are shown for two ligands with different binding affinities. Ligand binding is often characterized in terms of the concentration of ligand at which half of the receptor binding sites are occupied (k<sub>d</sub>). The ligand illustrated by the red curve has a higher binding affinity and smaller k<sub>d</sub> than the ligand illustrated by the green curve. If these two ligands were present at the same time, more of the higher affinity ligand would be bound to the available receptor binding sites. This is how [[carbon monoxide]] can compete |
+ | In the example shown to the left, ligand binding curves are shown for two ligands with different binding affinities. Ligand binding is often characterized in terms of the concentration of ligand at which half of the receptor binding sites are occupied, known as the [[dissociation constant]] (k<sub>d</sub>). The ligand illustrated by the red curve has a higher binding affinity and smaller k<sub>d</sub> than the ligand illustrated by the green curve. If these two ligands were present at the same time, more of the higher affinity ligand would be bound to the available receptor binding sites. This is how [[carbon monoxide]] can compete with [[oxygen]] in binding to hemoglobin, resulting in [[carbon monoxide poisoning]]. |
| + | |||
| + | Binding affinity is most commonly determined using a [[radiolabel]]ed ligand, known as hot ligand. ''Homologous competitive binding experiments'' involve binding-site competition between a hot ligand and a cold ligand (untagged ligand)<ref> |
||
| + | See |
||
| + | [http://www.graphpad.com/curvefit/homologous_.htm Homologous competitive binding curves], A complete guide to nonlinear regression, curvefit.com. |
||
| + | </ref>. |
||
| + | |||
| + | For the use of [[statistical mechanics]] in a quantitative study of the |
||
| + | ligand-receptor binding affinity, see the comprehensive article<ref> |
||
| + | Vu-Quoc, L., |
||
| + | [http://clesm.mae.ufl.edu/wiki.pub/index.php/Configuration_integral_%28statistical_mechanics%29 Configuration integral (statistical mechanics)], 2008. |
||
| + | </ref> |
||
| + | on the [[configuration integral]]. |
||
| + | |||
| + | ==Selective and non-selective== <!--non-selective redirects here--> |
||
| + | Selective ligands have a tendency to bind to a very limited types of receptors, while non-selective ligands bind to several types of receptors. This plays an important role in [[pharmacology]], where [[drugs]] that are non-selective tend to have more [[adverse effects]], because they bind to several other receptors in addition to the one generating the desired effect. |
||
== See also == |
== See also == |
||
* [[Allosteric regulation]] |
* [[Allosteric regulation]] |
||
| + | * [[Ki Database]] |
||
| + | * [[Schild regression]] |
||
| + | * [[Neural receptors]] |
||
==References== |
==References== |
||
| + | <references/> |
||
| + | |||
| + | ==Additional references== |
||
| + | |||
*[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Search&db=books&doptcmdl=GenBookHL&term=ligand+affinity+AND+endocrin%5Bbook%5D+AND+233023%5Buid%5D&rid=endocrin.section.33 Ligand binding to hormone receptors] in ''Endocrinology: An Integrated Approach'' by Stephen Nussey and Saffron Whitehead (2001) Published by BIOS Scientific Publishers Ltd. ISBN 1-85996-252-1. |
*[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Search&db=books&doptcmdl=GenBookHL&term=ligand+affinity+AND+endocrin%5Bbook%5D+AND+233023%5Buid%5D&rid=endocrin.section.33 Ligand binding to hormone receptors] in ''Endocrinology: An Integrated Approach'' by Stephen Nussey and Saffron Whitehead (2001) Published by BIOS Scientific Publishers Ltd. ISBN 1-85996-252-1. |
||
*[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Search&db=books&doptcmdl=GenBookHL&term=Molecular+Recognition+Processes+AND+cell%5Bbook%5D+AND+6191%5Buid%5D&rid=cell.section.298 Molecular Recognition Processes] in ''Molecular Biology of the Cell'' 3rd edition (1994) by Bruce Alberts, Dennis Bray, Julian Lewis, Martin Raff, Keith Roberts and James D. Watson. See Figure 3-9, [http://www.ncbi.nlm.nih.gov/books/bv.fcgi?call=bv.View..ShowSection&rid=cell.figgrp.316 Equilibrium ligand binding]. |
*[http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Search&db=books&doptcmdl=GenBookHL&term=Molecular+Recognition+Processes+AND+cell%5Bbook%5D+AND+6191%5Buid%5D&rid=cell.section.298 Molecular Recognition Processes] in ''Molecular Biology of the Cell'' 3rd edition (1994) by Bruce Alberts, Dennis Bray, Julian Lewis, Martin Raff, Keith Roberts and James D. Watson. See Figure 3-9, [http://www.ncbi.nlm.nih.gov/books/bv.fcgi?call=bv.View..ShowSection&rid=cell.figgrp.316 Equilibrium ligand binding]. |
||
| + | |||
| + | ==External links== |
||
| + | *[http://www.bindingdb.org BindingDB], a public database of measured protein-ligand binding affinities. |
||
{{Cell_signaling}} |
{{Cell_signaling}} |
||
| ⚫ | |||
| + | [[Category:Cell signaling]] |
||
| + | [[Category:Chemical bonding]] |
||
| + | [[Category:Neurochemistry]] |
||
| + | [[Category:Neurophysiology]] |
||
| ⚫ | |||
| + | [[Category:Ligands]] |
||
| + | <!-- |
||
| − | {{biochem-stub}} |
||
| + | [[da:Ligand (biokemi)]] |
||
| + | [[de:Ligand (Biochemie)]] |
||
| + | [[fr:Ligand (biologie)]] |
||
| + | [[it:Ligando (biochimica)]] |
||
| + | [[ja:リガンド]] |
||
| + | [[sv:Ligand (biokemi)]] |
||
| + | [[tr:Ligand (biyokimya)]] |
||
| + | [[zh:配體 (生物化學)]] |
||
| + | --< |
||
{{enWP|Ligand (biochemistry)}} |
{{enWP|Ligand (biochemistry)}} |
||
Latest revision as of 11:50, 7 March 2010
Assessment |
Biopsychology |
Comparative |
Cognitive |
Developmental |
Language |
Individual differences |
Personality |
Philosophy |
Social |
Methods |
Statistics |
Clinical |
Educational |
Industrial |
Professional items |
World psychology |
Biological: Behavioural genetics · Evolutionary psychology · Neuroanatomy · Neurochemistry · Neuroendocrinology · Neuroscience · Psychoneuroimmunology · Physiological Psychology · Psychopharmacology (Index, Outline)
Myoglobin (blue) with its ligand heme (orange) bound. Based on PDB ID 1MBO
Receptor binding or ligand binding is the process by which a ligand eg a neurotransmitter attaches itself to a receptor . In biochemistry, a ligand (latin ligare = to bind) is a substance that is able to bind to and form a complex with a biomolecule to serve a biological purpose. Such binding is specific, as in neurochemistry where neurotransmitters bind only to specific receptors so dopamine will bind to a dopamine receptor and not to any other.
In a narrower sense, it is a signal triggering molecule binding to a site on a target protein, by intermolecular forces such as ionic bonds, hydrogen bonds and Van der Waals forces. The docking (association) is usually reversible (dissociation). Actual irreversible covalent binding between a ligand and its target molecule is rare in biological systems. Ligand binding to receptors alters the chemical conformation, i.e. the three dimensional shape of the receptor protein. The conformational state of a receptor protein determines the functional state of a receptor. The tendency or strength of binding is called affinity. Ligands include substrates, inhibitors, activators, and neurotransmitters. Radioligands are radioisotope labeled compounds and used in vivo as tracers in PET studies and for in vitro binding studies.
Receptor/Ligand binding affinity
The interaction of most ligands with their binding sites can be characterized in terms of a binding affinity. In general, high affinity ligand binding results from greater intermolecular force between the ligand and its receptor while low affinity ligand binding involves less intermolecular force between the ligand and its receptor. In general, high affinity binding involves a longer residence time for the ligand at its receptor binding site than is the case for low affinity binding. High affinity binding of ligands to receptors is often physiologically important when some of the binding energy can be used to cause a conformational change in the receptor, resulting in altered behavior of an associated ion channel or enzyme.
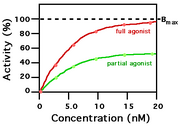
Two agonists with similar binding affinity
A ligand that can bind to a receptor, alter the function of the receptor and trigger a physiological response (eg the functioning of a neuron) is called an agonist for that receptor. Agonist binding to a receptor can be characterized both in terms of how much physiological response can be triggered and the concentration of the agonist that is required to produce the physiological response. High affinity ligand binding implies that a relatively low concentration of a ligand is adequate to maximally occupy a ligand binding site and trigger a physiological response. Low affinity binding implies that a relatively high concentration of a ligand is required before the binding site is maximally occupied and the maximum physiological response to the ligand is achieved. In the example shown to the right, two different ligands bind to the same receptor binding site. Only one of the agonists shown can maximally stimulate the receptor and thus can be defined as a "full agonist". An agonist that can only partially activate the physiological response is called a "partial agonist". Ligands that bind to a receptor but fail to activate the physiological response are receptor "antagonists". In this example, the concentration at which the full agonist (red curve) can half-maximally activate the receptor is about 5 x 10-9 Molar (nM = nanomolar).
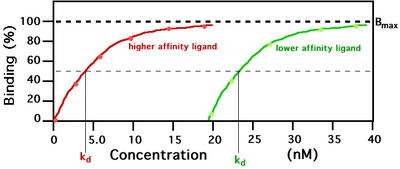
Two ligands with different receptor binding affinity.
In the example shown to the left, ligand binding curves are shown for two ligands with different binding affinities. Ligand binding is often characterized in terms of the concentration of ligand at which half of the receptor binding sites are occupied, known as the dissociation constant (kd). The ligand illustrated by the red curve has a higher binding affinity and smaller kd than the ligand illustrated by the green curve. If these two ligands were present at the same time, more of the higher affinity ligand would be bound to the available receptor binding sites. This is how carbon monoxide can compete with oxygen in binding to hemoglobin, resulting in carbon monoxide poisoning.
Binding affinity is most commonly determined using a radiolabeled ligand, known as hot ligand. Homologous competitive binding experiments involve binding-site competition between a hot ligand and a cold ligand (untagged ligand)[1].
For the use of statistical mechanics in a quantitative study of the ligand-receptor binding affinity, see the comprehensive article[2] on the configuration integral.
Selective and non-selective
Selective ligands have a tendency to bind to a very limited types of receptors, while non-selective ligands bind to several types of receptors. This plays an important role in pharmacology, where drugs that are non-selective tend to have more adverse effects, because they bind to several other receptors in addition to the one generating the desired effect.
See also
- Allosteric regulation
- Ki Database
- Schild regression
- Neural receptors
References
- ↑ See Homologous competitive binding curves, A complete guide to nonlinear regression, curvefit.com.
- ↑ Vu-Quoc, L., Configuration integral (statistical mechanics), 2008.
Additional references
- Ligand binding to hormone receptors in Endocrinology: An Integrated Approach by Stephen Nussey and Saffron Whitehead (2001) Published by BIOS Scientific Publishers Ltd. ISBN 1-85996-252-1.
- Molecular Recognition Processes in Molecular Biology of the Cell 3rd edition (1994) by Bruce Alberts, Dennis Bray, Julian Lewis, Martin Raff, Keith Roberts and James D. Watson. See Figure 3-9, Equilibrium ligand binding.
External links
- BindingDB, a public database of measured protein-ligand binding affinities.
Cell physiology: cell signaling | |
|---|---|
| Key concepts |
Ligand - Cell signaling networks - Signal transduction - Apoptosis - Second messenger system (Ca2+ signaling, Lipid signaling) |
| Processes |
Paracrine - Autocrine - Juxtacrine - Neurotransmitters - Endocrine (Neuroendocrine) |
| Types of proteins |
Receptor (Transmembrane, Intracellular) - Transcription factor (General, Preinitiation complex, TFIID, TFIIH) - Adaptor protein |