| Line 67: | Line 67: | ||
*[http://www.compchemwiki.org/index.php?title=Purine Computational Chemistry Wiki] |
*[http://www.compchemwiki.org/index.php?title=Purine Computational Chemistry Wiki] |
||
| − | [[Category:Purines]] |
+ | [[Category:Purines|*]] |
[[Category:Biochemicals]] |
[[Category:Biochemicals]] |
||
Latest revision as of 23:28, 11 August 2006
Assessment |
Biopsychology |
Comparative |
Cognitive |
Developmental |
Language |
Individual differences |
Personality |
Philosophy |
Social |
Methods |
Statistics |
Clinical |
Educational |
Industrial |
Professional items |
World psychology |
Biological: Behavioural genetics · Evolutionary psychology · Neuroanatomy · Neurochemistry · Neuroendocrinology · Neuroscience · Psychoneuroimmunology · Physiological Psychology · Psychopharmacology (Index, Outline)
| Purine | |
|---|---|
| Chemical name | Purine |
| Chemical formula | C5H4N4 |
| Molecular mass | 120.11 g/mol |
| Melting point | 214 °C |
| CAS number | 120-73-0 |
| SMILES | C1(NC=N2)=C2C=NC=N1 |
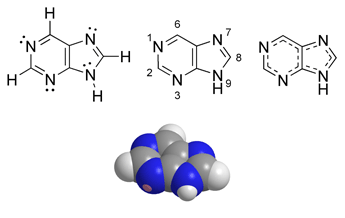 | |
Purine is a heterocyclic aromatic organic compound, consisting of a pyrimidine ring fused to an imidazole ring.
The general term purines also refers to substituted purines and their tautomers. Two of the bases in nucleic acids, adenine and guanine, are purines. In DNA, these bases form hydrogen bonds with their complementary pyrimidines thymine and cytosine.
In RNA, the complement of adenine is uracil (U) instead of thymine.
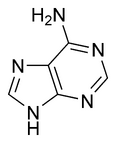
|

|
| Adenine | Guanine |
These hydrogen bonding modes are for classical Watson-Crick base pairing. Other hydrogen bonding modes are seen in both DNA and RNA. Of significance, the additional 2'-hydroxyl group of the ribose moiety in RNA expands the configurations through which RNA can form hydrogen bonds.
Other notable purines are xanthine, hypoxanthine, theobromine, caffeine and uric acid.
Purines are biochemically significant as components of DNA and RNA, and are also found in a number of other important biomolecules, such as ATP, GTP, cyclic AMP, NADH, and coenzyme A.
Metabolism
Many organisms have metabolic pathways to synthesise and break down purines.
Purines are biologically synthesised as nucleosides (bases attached to ribose). Both adenine and guanine are derived from the nucleoside inosine monophosphate, which is synthesised on a pre-existing ribose through a complex pathway using atoms from the amino acids glycine, glutamine, and aspartic acid, as well as formate ions transferred from the coenzyme tetrahydrofolate.
Purines from food (or from tissue turnover) are metabolised by several enzymes, including xanthine oxidase, into uric acid. High levels of uric acid can predispose to gout when the acid crystalises in joints; this phenomenon only happens in humans and some animal species (e.g. dogs) that lack an intrinsic uricase enzyme that can further degrade uric acid. The deficiency of another enzyme, adenosine deaminase, needed to break down adenine, is a cause of severe combined immunodeficiency.
Purines from turnover of nucleic acids (or from food) can also be salvaged and reused in new nucleotides. The enzyme adenine phosphoribosyltransferase salvages adenine, while hypoxanthine-guanine phosphoribosyltransferase (HPRT) salvages guanine and hypoxanthine. Genetic deficiency of HPRT causes Lesch-Nyhan syndrome.
History
Purine was named by the German chemist Emil Fischer in 1884. He synthesised it in 1898. Fischer showed that the purines were part of a single chemical family.
See also
- Simple aromatic rings
External links
| This page uses Creative Commons Licensed content from Wikipedia (view authors). |
