In genetics, splicing is a modification of genetic information after transcription, in which introns are removed and exons are joined. Splicing prepares precursor messenger RNA in eukaryotes to produce mature messenger RNA. This mature messenger RNA is then prepared to undergo translation as part of protein synthesis to produce proteins. Splicing occurs by a series of biochemical reactions between RNA nucleotides, which are catalyzed by proteins, RNA, or both.

Simple illustration of exons and introns in pre-mRNA. The mature mRNA is formed by splicing.
Splicing pathways[]
Several methods of RNA splicing occur in nature. The type of splicing depends on the structure of the spliced intron and the catalysts required for splicing to occur. Regardless of which pathway is used, the excised introns are discarded.
Spliceosomal[]
Spliceosomal introns often reside in eukaryotic protein-coding genes. Within the intron, a 3' splice site, 5' splice site, and branch site are required for splicing. Splicing is catalyzed by the spliceosome which is a large RNA-protein complex composed of five small nuclear ribonucleoproteins (snRNPs, pronounced "snurps"). The RNA components of snRNPs interact with the intron and may be involved in catalysis. Two types of spliceosomes have been identified (the major and minor) which contain different snRNPs.
- Major
- The major spliceosome splices introns containing GU at the 5' splice site and AG at the 3' splice site. It is composed of the U1, U2, U4, U5, and U6 snRNPs.
U1- binds 5' splice site U2- binds the branch U4- inhibits U6, lost to activate spliceosome U5 - binds U1 and U2 to create lariat U6 - When, activated, displaces U1 and binds U2. U2-U6 forms active catalytic complex
- Minor
- The minor spliceosome is very similar to the major spliceosome, however it splices rare introns with different splice site sequences. Here, the 3' and 5' splice sites are AU and AC, respectively. While the minor and major spliceosomes contain the same U5 snRNP, the minor spliceosome has different, but functionally analogous snRNPs for U1, U2, U4, and U6, which are respectively called U11, U12, U4atac, and U6atac. [1]
- Trans-splicing
- Trans-splicing is a form of splicing that joins two exons that are not within the same RNA transcript.
Self-splicing[]
Self-splicing occurs for rare introns that form a ribozyme, performing the functions of the spliceosome by RNA alone. There are three kinds of self-splicing introns, Group I, II, and III. Group II and III introns perform splicing similar to the spliceosome without requiring any protein. This similarity suggests that Group II and III introns may be evolutionarily related to the spliceosome. Self-splicing may also be very ancient, and may have existed in an RNA world that was present before protein.
tRNA splicing[]
tRNA (also tRNA-like) splicing is another rare form of splicing that usually occurs in tRNA. The splicing reaction involves a different biochemistry than the spliceomsomal and self-splicing pathways. Ribonucleases cleave the RNA and ligases join the exons together. This form of splicing does also not require any RNA components for catalysis.
Evolution[]
Splicing occurs in all the kingdoms or domains of life, however, the extent and types of splicing can be very different between the major divisions. Eukaryotes splice many protein-coding messenger RNAs and some non-coding RNAs. Prokaryotes, on the other hand, splice rarely, but mostly non-coding RNAs. Another important difference between these two groups of organisms is that prokaryotes completely lack the spliceosomal pathway.
Because spliceosomal introns are not conserved in all species, there is debate concerning when spliceosomal splicing evolved. Two models have been proposed: the intron late and intron early models (see intron evolution).
| Eukaryotes | Prokaryotes | |
|---|---|---|
| Spliceosomal | + | - |
| Self-splicing | + | + |
| tRNA | + | + |
Biochemical mechanism[]
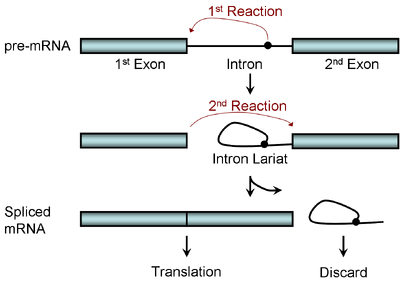
Diagram illustrating the two-step biochemistry of splicing
Spliceosomal splicing and self-splicing involves a two-step biochemical process. Both steps involve transesterification reactions that occur between RNA nucleotides. tRNA splicing, however, is an exception and does not occur by transesterification.
Spliceosomal and self-splicing transesterification reactions occur in a specific order. First, a specific branch-point nucleotide within the intron reacts with the first nucleotide of the intron, forming an intron lariat. Second, the last nucleotide of the first exon reacts with the first nucleotide of the second exon, joining the exons and releasing the intron lariat.
Computer programs to find splicing sites in genomic DNA sequences[]
Alternative splicing[]
Main article: Alternative splicing
In many cases, the splicing process can create many unique proteins by variations in the splicing of the same messenger RNA. This phenomenon is called alternative splicing.
Experimental manipulation of splicing[]
Splicing events can be experimentally altered[2], [3] by binding steric-blocking antisense oligos such as Morpholinos or PNAs to snRNP binding sites, to the branchpoint nucleotide that closes the lariat, or to splice-regulatory element binding sites[4].
Splicing errors[]
Mutations in the introns or exons can prevent splicing and thus may prevent protein biosynthesis.
Common errors:
- Mutation of a splice site resulting in loss of function of that site. Results in exposure of a premature stop codon, loss of an exon, or inclusion of an intron.
- Mutation of a splice site reducing specificity. May result in variation in the splice location, causing insertion or deletion of amino acids, or most likely, a loss of the reading frame.
- Transposition of a splice site, resulting in inclusion or excluson of more DNA than expected. Results in longer or shorter exons.
References[]
1. Patel, A.A. and J.A. Steitz, Splicing Double: Insights from the Second Spliceosome. Nature Reviews Molecular Cell Biology, 2003. 4(12): p. 960.
2. Draper BW, Morcos PA, Kimmel CB. Inhibition of zebrafish fgf8 pre-mRNA splicing with morpholino oligos: A quantifiable method for gene knockdown. Genesis. 2001 Jul;30(3):154-6.
3. Sazani P, Kang SH, Maier MA, Wei C, Dillman J, Summerton J, Manoharan M, Kole R. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res. 2001 Oct 1;29(19):3965-74.
4. Bruno IG, Jin W, Cote GJ. Correction of aberrant FGFR1 alternative RNA splicing through targeting of intronic regulatory elements. Hum Mol Genet. 2004 Oct 15;13(20):2409-20. Epub 2004 Aug 27.
See also[]
| Post Transcriptional Modification |
|---|
|
Transcription | Post transcriptional modification | RNA splicing | Polyadenylation | 5' cap |
de:Spleißen (Biologie) es:Ayuste eo:Splisado fr:Épissage he:שחבור (ביולוגיה) nl:Splicing pt:Splicing fi:Silmukointi
| This page uses Creative Commons Licensed content from Wikipedia (view authors). |